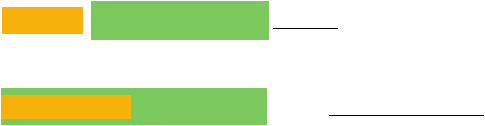
I. Fluorescent tags: Citrine-YFP and CFP
We routinely tag proteins with the Citrine variant of Yellow Fluorescent Protein (YFP)
(Griesbeck et al. 2001), which can be used not only to visualize a single protein but also to study
protein-protein interactions in vivo as an energy acceptor in BRET (Xu et al. 1999) and FRET assays
(Tsien et al. 1998; Pollok et al. 1999). Moreover, Citrine-YFP has enhanced photostability and is much
less sensitive to pH and anions, such as chloride, compared to other YFP variants (Griesbeck et al.
2001). The reduced sensitivity to pH allows detection of proteins targeted to the extracellular matrix or
to other relatively acidic subcellular compartments, thus making this reporter more suitable for tagging
proteins with a wide range of targeting specificities. Some proteins are also tagged with Cyan
Fluorescent Protein (CFP) (ECFP, Clontech) for comparison of localization patterns obtained with
different tags and for future colocalization and interaction studies.
For tagging, Citrine-YFP/CFP coding sequences, which lack start and stop codons, are flanked
by linker peptides that function as flexible tethers, minimizing potential folding interference between
Citrine-YFP/CFP and the tagged protein (Doyle et al. 1996). To avoid placing identical nucleotide
sequences on each side of the tag, we use two different linkers: the N-terminus of the tag is linked to a
glycine-rich linker peptide (Gly)
5
Ala, and the C-terminus is linked to an alanine-rich linker peptide
AlaGly(Ala)
5
GlyAla.
FseI
SfiI
AlaGly(Ala)
5
GlyAla
(Gly)
5
Ala
Citrine-YFP/CFP C-terminus
Citrine-YFP/CFP N-terminus
Figure 1
. Forward and reverse primers for adding flanking linkers and restriction sites to citrine-YFP/CFP.
Orange boxes indicate the FseI and SfiI sites in the forward and reverse primers, respectively. Green
boxes indicate the (Gly)
5
Ala and AlaGly
(Ala)
5
GlyAla linkers in the forward and reverse primers,
respectively. N-terminal and C-terminal sequences of Citrine-YFP/CFP contained in the forward and
reverse primers, respectively, are indicated in blue.
forward primer FseI
-Gly
-Citrine-YFP/CFP
5’-AA GGC CGG CCT GGA GGT GGA GGT GGA GCT
GTG AGC A
-3’
reverse primer SfiI-Ala-Citrine-YFP/CFP
5’-TT GGC CCC AGC GGC CGC AGC AGC ACC AGC AGG ATC
CTT GTA CAG CTC GTC CA
-3’
The Citrine-YFP tag is amplified from the pRSET
B
-Citrine plasmid (Griesbeck et al. 2001) and
the CFP tag from the pECFP-C1 plasmid (Clontech) using the ExTaq DNA polymerase (TaKaRa) and
two primers shown in Figure 1. The products are cloned into the pTOPO TA vector (Invitrogen). The
resulting cDNAs encoding the fluorescent tags contain recognition sequences for FseI and SfiI
restriction endonucleases at their 5’- and 3’-ends, respectively. Plasmid with Citrine-YFP is designated
as pCitrine-3 and plasmid with CFP as pCFP-3 (Figure 2).


