4
b. Primers
Two sets of primers (P1/P2, P3/P4) for each gene are designed for the amplification of two
genomic fragments using the Primer3 software (http://www-genome.wi.mit.edu/cgi-
bin/primer/primer3_www.cgi). Our PCR design program considers a series of criteria including the
position of each primer within the genomic sequence, annealing temperature, length, and hairpin
structures in an iterative fashion to determine the most suitable sets of P1/P2 and P3/P4 for each gene.
The first set of primers amplifies a fragment (P1-P2) that extends from up to 3 kb upstream of
the transcription start of the gene to the tag insertion site within the coding sequence. We believe that
most Arabidopsis promoters should be contained within 3 kb. However, some intergenic regions are <3
kb; thus, we defined a minimal size for the 5’ UTR and promoter region as 1 kb, extending P1 into the
upstream ORF if the intergenic region is very small.
The second set of primers amplifies a fragment (P3-P4) from the tag insertion site to 0.5-1 kb
downstream of the gene to include 3’ UTR and regulatory sequences. The default position for the start
of the gene-specific region of P2 and P3 primers is at the 30th nucleotide (i.e. 10 amino acids) upstream
of the stop codon. However, if a functional domain is predicted at this position, or it does not generate
a suitable primer sequence, the positions of P2 and P3 are reiteratively shifted from the initial site until
suitable priming sites are determined.
P1 and P4 contain, in addition to gene-specific sequences, sequences partially overlapping the
attB1 and attB2 Gateway forward and reverse primers, respectively (used for TT-PCR, see below). P2
and P3 contain sequences partially overlapping the Citrine-YFP/CFP primers (Figure 5).
P1 primer: 5’-GCTCGATCCACCTAGGCT
+18-25 gene-specific nucleotides-3’
P2 primer: 5’-CACAGCTCCACCTCCACCTCCAGGCCGGCC
+18-25 gene-specific nucleotides-3’
P3 primer: 5’-TGCTGGTGCTGCTGCGGCCGCTGGGGCC
+18-25 gene-specific nucleotides-3’
P4 primer: 5’-CGTAGCGAGACCACAGGA
+18-25 gene-specific nucleotides-3’
Figure 5
. Nucleotide sequences of P1, P2, P3, and P4 primers. Gene-non-specific sequences overlapping forwar
attB1 and reverse attB2 Gateway primers are indicated in dark red, and gene-non-specific sequences overlapping
the Citrine-YFP/CFP primers are indicated in blue.
c. PCR conditions
Because all primers in the TT-PCR reaction (P1-P4) have gene-specific sequences, it is
impossible to calculate a standard annealing temperature for all genes to be tagged. Instead, we use
touch-down PCR conditions to include a range of temperatures as shown below.
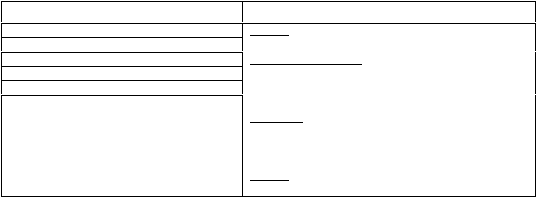
PCR reaction mixture
PCR cycles
100 ng DNA template
1x ExTaq reaction buffer
0.2 mM 4 x dNTP
0.2 µM of each primer
0.025 U/µl ExTaq (TaKaRa)
total volume: 20 µl
1 cycle:
95°C
2 min 30 sec
7 cycles touch-down:
94°C
30 sec
64°C
30 sec; reduce t°C by 1°C per cycle
68°C
1 min per kb
23 cycles:
94°C
30 sec
58°C
30 sec
68°C
1 min per kb
1 cycle:
68°C
10 min

